Transcription Factor Footprinting
Learning outcomes
detect transcription factor binding signatures in ATAC-seq data
|
Introduction
While ATAC-seq can uncover accessible regions where transcription factors (TFs) might bind, reliable identification of specific TF binding sites (TFBS) still relies on chromatin immunoprecipitation methods such as ChIP-seq. ChIP-seq methods require high input cell numbers, are limited to one TF per assay, and are further restricted to TFs for which antibodies are available. Therefore, it remains costly, or even impossible, to study the binding of multiple TFs in one experiment.
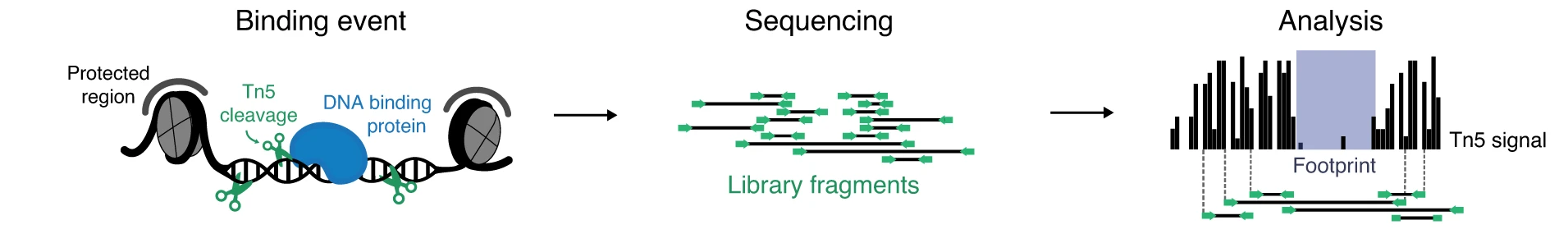
Similarly to nucleosomes, bound TFs hinder cleavage of DNA, resulting in footprints: defined regions of decreased signal strength within larger regions of high signal (Figure 1.).
Despite its compelling potential, a number of issues have rendered footprinting of ATAC-seq data cumbersome. It has been described that enzymes used in chromatin accessibility assays (e.g., DNase-I, Tn5) are biased towards certain sequence compositions. If unaccounted for, this impairs the discovery of true TF footprints.
In this tutorial we use an R / Bioconductor packages ATACseqQC and MotifDb to detect TF binding signatures in ATAC-seq data. Please note this tutorial is merely an early attempt to determine whether TF bindng sites can be identified in ATAC-seq data, and not a statistical framework for TF footrpinting. It can be used as a QC step to detect expected TF sites in the data rather than for discovery of novel binding patterns.
Data & Methods
We will build upon the lab ATACseq specifc QC and use the same data as for other ATAC-seq labs.
Setting-up
We need access to bam file with shifted alignments shifted.bam which we created in the ATACseq specifc QC. This file contains alignments shifted +4 bps on the + strand and -5 bps on the - strand, to account for Tn5 transposition.
Assuming we start at analysis:
mkdir TF_footprnt
cd TF_footprnt
ln -s ../QC/splitBam/shifted.bam .
ln -s ../QC/splitBam/shifted.bam.bai .
module load R_packages/4.1.1
Hint
Please check first that file shifted.bam exists in this location: ls ../QC/splitBam/shifted.bam. If it does, the output of this command is the path; if it does not you get “file does not exist”. You can link the file prepared earlier:
ln -s ../../results/QC/splitBam/shifted.bam
ln -s ../../results/QC/splitBam/shifted.bam.bai
We activate R console upon typing R in the terminal.
Detection of TF Binding Signatures
We begin by loading necessary libraries:
library(ATACseqQC)
library(MotifDb)
library(BSgenome.Hsapiens.UCSC.hg38)
genome <- Hsapiens
We load data from shifted bam file:
shifted.bamFile="shifted.bam"
# we will limit the analysis to chr14
seqlev <- "chr14"
Let’s first check signatures for a general TF CTCF. This is its motif as position weight matrix (PWM), which consists of frequencies of each base at each motif position:
CTCF <- query(MotifDb, c("CTCF"))
CTCF <- as.list(CTCF)
print(CTCF[[1]], digits=2)
CTCF PWM
CTCF PWM:
1 2 3 4 5 6 7 8 9 10 11 12 13
A 0.10 0.16 0.30 0.072 0.012 0.786 0.024 0.122 0.914 0.012 0.376 0.022 0.028
C 0.36 0.21 0.10 0.826 0.966 0.024 0.620 0.494 0.010 0.008 0.010 0.022 0.002
G 0.12 0.41 0.44 0.050 0.012 0.108 0.336 0.056 0.048 0.976 0.602 0.606 0.962
T 0.42 0.22 0.16 0.052 0.010 0.082 0.020 0.328 0.028 0.004 0.012 0.350 0.008
14 15 16 17 18 19
A 0.024 0.096 0.424 0.086 0.12 0.34
C 0.016 0.818 0.024 0.532 0.35 0.26
G 0.880 0.038 0.522 0.326 0.12 0.32
T 0.080 0.048 0.030 0.056 0.41 0.08
We now summarise the signal in the vicinity of CTCF motifs (100 bps up- and down-stream):
ctcf <- factorFootprints(shifted.bamFile, pfm=CTCF[[1]],
genome=genome,
min.score="90%", seqlev=seqlev,
upstream=100, downstream=100)
This function outputs signal mean values of coverage for positive strand and negative strand in feature regions, and other information which you can inspect using str(ctcf):
spearman.correlationspearman correlations of cleavage counts in the highest 10-nucleotide-windowbindingSites- GRanges object with detected bindng sitesProfile.segmentation
Let’s inspect the statistics ctcf$spearman.correlation:
> ctcf$spearman.correlation
$`+`
Spearman's rank correlation rho
data: predictedBindingSiteScore and highest.sig.windows
S = 3761558200, p-value < 2.2e-16
alternative hypothesis: true rho is not equal to 0
sample estimates:
rho
0.2653951
$`-`
Spearman's rank correlation rho
data: predictedBindingSiteScore and highest.sig.windows
S = 3801340300, p-value < 2.2e-16
alternative hypothesis: true rho is not equal to 0
sample estimates:
rho
0.2576259
The plot produced by this function is of signal mean values of coverage for positive strand and negative strand in feature regions.
pdf("ctcf_footprnt.pdf")
sigs <- factorFootprints(shifted.bamFile, pfm=CTCF[[1]],
genome=genome,
min.score="90%", seqlev=seqlev,
upstream=100, downstream=100)
dev.off()
We can generate similar plots for other TFs.
RFX5
RFX5 <- query(MotifDb, c("RFX5"))
RFX5 <- as.list(RFX5)
rfx5 <- factorFootprints(shifted.bamFile, pfm=RFX5[[1]],
genome=genome,
min.score="90%", seqlev=seqlev,
upstream=100, downstream=100)
rfx5$spearman.correlation$`+`$estimate
rfx5$spearman.correlation$`+`$p.value
pdf("rfx5_footprnt.pdf")
rfx5 <- factorFootprints(shifted.bamFile, pfm=RFX5[[1]],
genome=genome,
min.score="90%", seqlev=seqlev,
upstream=100, downstream=100)
dev.off()
STAT3
STAT3 <- query(MotifDb, c("STAT3"))
STAT3 <- as.list(STAT3)
stat3 <- factorFootprints(shifted.bamFile, pfm=STAT3[[1]],
genome=genome,
min.score="90%", seqlev=seqlev,
upstream=100, downstream=100)
stat3$spearman.correlation$`+`$estimate
stat3$spearman.correlation$`+`$p.value
stat3$Profile.segmentation
pdf("stat3_footprnt.pdf")
stat3 <- factorFootprints(shifted.bamFile, pfm=STAT3[[1]],
genome=genome,
min.score="90%", seqlev=seqlev,
upstream=100, downstream=100)
dev.off()
Which factors show evidence of binding enrichment in this data set?
CTCF |
RFX5 |
STAT3 |
|---|---|---|
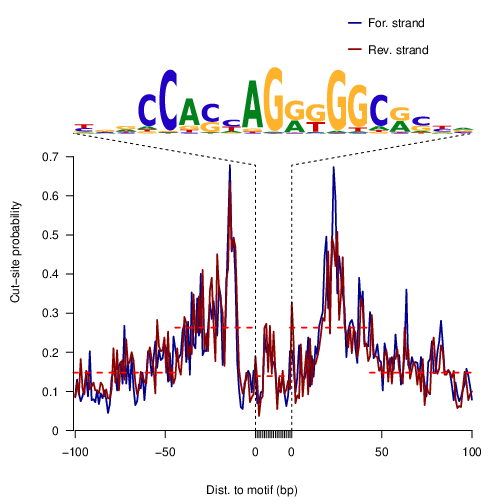
|
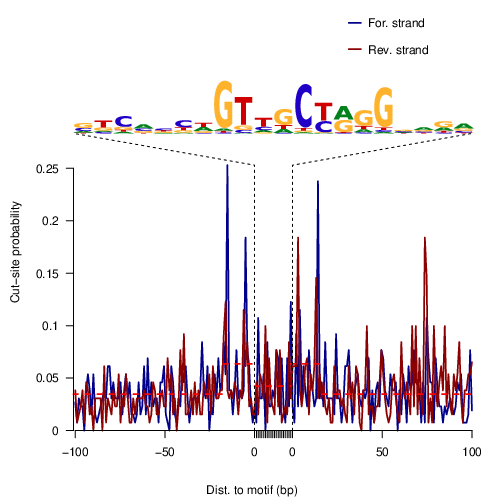
|
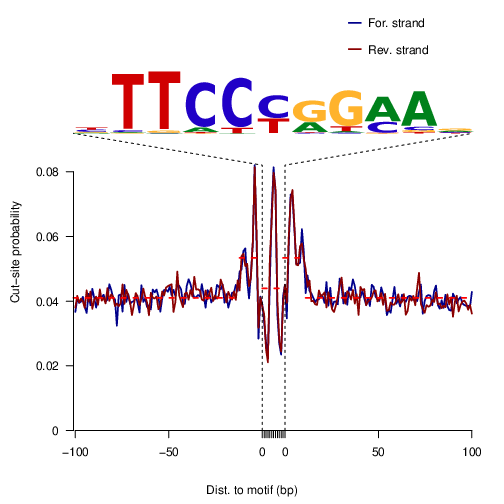
|
image source: https://doi.org/10.1038/s41467-020-18035-1 (Figure 1.)